The genotype-phenotype correlation in geographic atrophy

Age-related macular degeneration (AMD) is a disease that advances in successive phases, in which drusen (identified as yellowish deposits in the retinography located between the Bruch’s membrane and the retinal pigment epithelium -RPE) are the common link. Some patients remain in the initial phases of AMD for years without major vision loss, while others progress quickly to the most advanced phases, in which vision worsens markedly. The two advanced types are neovascular AMD and geographic atrophy (GA). The latter is believed to represent the end of the natural progression of the disease if neovascular AMD does not appear.
GA is diagnosed in a patient of over 50 when there is identification of a loss of RPE, adjacent photoreceptors and choriocapillaris, accompanied by drusen. Unfortunately, GA is untreatable today. In addition, a common characteristic in GA is the progressive growth of the atrophic area, with a speed that varies greatly between patients. Even though the specific causes of AMD are unknown, in recent years there have been major advances produced, among others, by the discovery of over 50 genetic variants (called Single Nucleotide Polymorphisms (SNP)) associated with AMD (1). These genes have implied the activity of the Complement pathway (which forms part of the innate immune system), the lipid metabolism and the extracellular matrix in the pathogenesis of the disease.

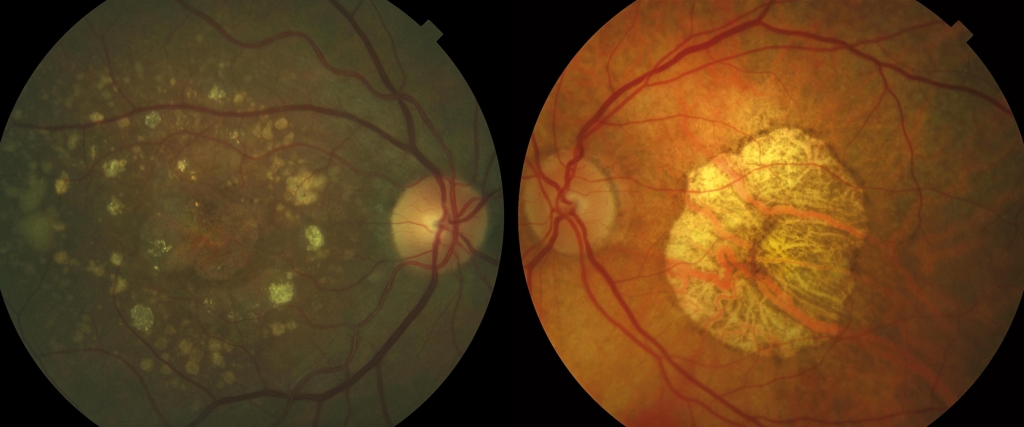
Driven by the variety of phenotypes (aspect of the eye fundus) displayed by patients with GA, researchers from the Barcelona Macula Foundation: Research for Vision (BMF) published a study in which, using the statistical technique called cluster analysis, they identified three groups of patients with GA (2). The first group was characterised by the presence of foveal atrophy and a large number of soft drusen, another group by the presence of reticular drusen, perifoveal atrophy and a reduced thickness of the choroid (the vascular layer adjacent to the retina), and a third group with intermediate characteristics. In addition, the progression of the RPE atrophy varied between groups, with a markedly slower growth in the first. Therefore, it seems that the term “geographic atrophy” could embrace disease subtypes with different prognoses (see photo below).
The following step is to determine whether these groups correspond to different genetic variants. Put differently: could the genetic profile of a patient determine, not only the aspect of GA, but also the aspect of the eye fundus (the phenotype)? This could help to determine a patient’s genetic alterations that are associated with the disease in order to better understand the mechanism by which these lesions occur and, in the future, to the development of rational therapies for each. In fact, there are precedents for this being the case in AMD, where patients with certain characteristics of the eye fundus have been associated with certain genetic variants (3-4).
This is precisely one of the studies in which BMF researchers are participating: EYE-RISK, a multicentric, multidisciplinary European project that includes dozens of researchers from different countries in the continent. The teams of Queen’s University Belfast and the BMF will assess the possible relationship between the genotype and phenotype of the patients included in AMD studies in Europe by using an innovative methodology based on Deep Learning (a type of Artificial Intelligence). If these hypotheses are confirmed, the development of treatments would have to correct the specific mechanisms by which the disease develops in each. This is known as “personalised medicine” or “precision medicine”: the right treatment for the right patient at the right time.
Author: Marc Biarnés
Bibliographic references:
1. Fritsche LG i cols. A genome-wide association study of age-related macular degeneration highlights contributions of rare and common variants.
Nat Genet 2016; 48: 134-43.
2. Monés J i cols. Geographic atrophy phenotype identification by cluster analysis. Br J Ophthalmol 2018; 102: 388-92.
3. van Asten F i cols. A deep phenotype association study reveals specific phenotype associations wih genetic variants in age-related macular degeneration. Opthalmology 2018; 125: 559-68.
4. Kersten E I cols. Phenotype characteristics of patients with age-related macular degeneration carrying a rare variant in the Complement Factor H gene. JAMA Ophthalmol 2017; 135: 1037-44.